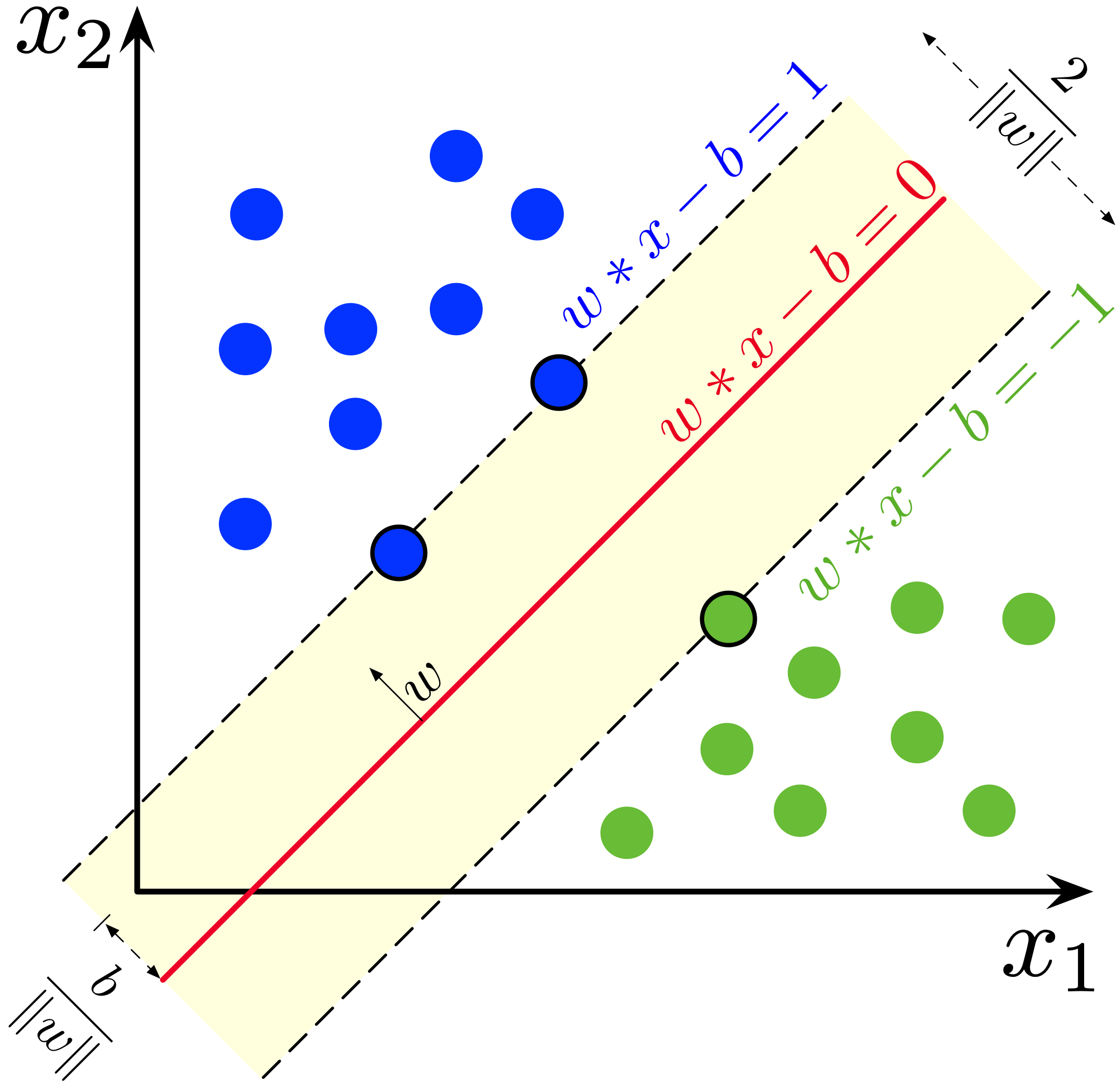
Fitting 5 folds for each of 25 candidates, totalling 125 fits
[CV 1/5] END .................................C=0.1, gamma=1; total time= 0.1s
[CV 2/5] END .................................C=0.1, gamma=1; total time= 0.0s
[CV 3/5] END .................................C=0.1, gamma=1; total time= 0.0s
[CV 4/5] END .................................C=0.1, gamma=1; total time= 0.0s
[CV 5/5] END .................................C=0.1, gamma=1; total time= 0.0s
[CV 1/5] END ...............................C=0.1, gamma=0.1; total time= 0.0s
[CV 2/5] END ...............................C=0.1, gamma=0.1; total time= 0.0s
[CV 3/5] END ...............................C=0.1, gamma=0.1; total time= 0.0s
[CV 4/5] END ...............................C=0.1, gamma=0.1; total time= 0.0s
[CV 5/5] END ...............................C=0.1, gamma=0.1; total time= 0.0s
[CV 1/5] END ..............................C=0.1, gamma=0.01; total time= 0.0s
[CV 2/5] END ..............................C=0.1, gamma=0.01; total time= 0.0s
[CV 3/5] END ..............................C=0.1, gamma=0.01; total time= 0.0s
[CV 4/5] END ..............................C=0.1, gamma=0.01; total time= 0.0s
[CV 5/5] END ..............................C=0.1, gamma=0.01; total time= 0.0s
[CV 1/5] END .............................C=0.1, gamma=0.001; total time= 0.0s
[CV 2/5] END .............................C=0.1, gamma=0.001; total time= 0.0s
[CV 3/5] END .............................C=0.1, gamma=0.001; total time= 0.0s
[CV 4/5] END .............................C=0.1, gamma=0.001; total time= 0.0s
[CV 5/5] END .............................C=0.1, gamma=0.001; total time= 0.0s
[CV 1/5] END ............................C=0.1, gamma=0.0001; total time= 0.0s
[CV 2/5] END ............................C=0.1, gamma=0.0001; total time= 0.0s
[CV 3/5] END ............................C=0.1, gamma=0.0001; total time= 0.0s
[CV 4/5] END ............................C=0.1, gamma=0.0001; total time= 0.0s
[CV 5/5] END ............................C=0.1, gamma=0.0001; total time= 0.0s
[CV 1/5] END ...................................C=1, gamma=1; total time= 0.0s
[CV 2/5] END ...................................C=1, gamma=1; total time= 0.0s
[CV 3/5] END ...................................C=1, gamma=1; total time= 0.0s
[CV 4/5] END ...................................C=1, gamma=1; total time= 0.0s
[CV 5/5] END ...................................C=1, gamma=1; total time= 0.0s
[CV 1/5] END .................................C=1, gamma=0.1; total time= 0.0s
[CV 2/5] END .................................C=1, gamma=0.1; total time= 0.0s
[CV 3/5] END .................................C=1, gamma=0.1; total time= 0.0s
[CV 4/5] END .................................C=1, gamma=0.1; total time= 0.0s
[CV 5/5] END .................................C=1, gamma=0.1; total time= 0.0s
[CV 1/5] END ................................C=1, gamma=0.01; total time= 0.0s
[CV 2/5] END ................................C=1, gamma=0.01; total time= 0.0s
[CV 3/5] END ................................C=1, gamma=0.01; total time= 0.0s
[CV 4/5] END ................................C=1, gamma=0.01; total time= 0.0s
[CV 5/5] END ................................C=1, gamma=0.01; total time= 0.0s
[CV 1/5] END ...............................C=1, gamma=0.001; total time= 0.0s
[CV 2/5] END ...............................C=1, gamma=0.001; total time= 0.0s
[CV 3/5] END ...............................C=1, gamma=0.001; total time= 0.0s
[CV 4/5] END ...............................C=1, gamma=0.001; total time= 0.0s
[CV 5/5] END ...............................C=1, gamma=0.001; total time= 0.0s
[CV 1/5] END ..............................C=1, gamma=0.0001; total time= 0.0s
[CV 2/5] END ..............................C=1, gamma=0.0001; total time= 0.0s
[CV 3/5] END ..............................C=1, gamma=0.0001; total time= 0.0s
[CV 4/5] END ..............................C=1, gamma=0.0001; total time= 0.0s
[CV 5/5] END ..............................C=1, gamma=0.0001; total time= 0.0s
[CV 1/5] END ..................................C=10, gamma=1; total time= 0.0s
[CV 2/5] END ..................................C=10, gamma=1; total time= 0.0s
[CV 3/5] END ..................................C=10, gamma=1; total time= 0.0s
[CV 4/5] END ..................................C=10, gamma=1; total time= 0.0s
[CV 5/5] END ..................................C=10, gamma=1; total time= 0.0s
[CV 1/5] END ................................C=10, gamma=0.1; total time= 0.0s
[CV 2/5] END ................................C=10, gamma=0.1; total time= 0.0s
[CV 3/5] END ................................C=10, gamma=0.1; total time= 0.0s
[CV 4/5] END ................................C=10, gamma=0.1; total time= 0.0s
[CV 5/5] END ................................C=10, gamma=0.1; total time= 0.0s
[CV 1/5] END ...............................C=10, gamma=0.01; total time= 0.0s
[CV 2/5] END ...............................C=10, gamma=0.01; total time= 0.0s
[CV 3/5] END ...............................C=10, gamma=0.01; total time= 0.0s
[CV 4/5] END ...............................C=10, gamma=0.01; total time= 0.0s
[CV 5/5] END ...............................C=10, gamma=0.01; total time= 0.0s
[CV 1/5] END ..............................C=10, gamma=0.001; total time= 0.0s
[CV 2/5] END ..............................C=10, gamma=0.001; total time= 0.0s
[CV 3/5] END ..............................C=10, gamma=0.001; total time= 0.0s
[CV 4/5] END ..............................C=10, gamma=0.001; total time= 0.0s
[CV 5/5] END ..............................C=10, gamma=0.001; total time= 0.0s
[CV 1/5] END .............................C=10, gamma=0.0001; total time= 0.0s
[CV 2/5] END .............................C=10, gamma=0.0001; total time= 0.0s
[CV 3/5] END .............................C=10, gamma=0.0001; total time= 0.0s
[CV 4/5] END .............................C=10, gamma=0.0001; total time= 0.0s
[CV 5/5] END .............................C=10, gamma=0.0001; total time= 0.0s
[CV 1/5] END .................................C=100, gamma=1; total time= 0.0s
[CV 2/5] END .................................C=100, gamma=1; total time= 0.0s
[CV 3/5] END .................................C=100, gamma=1; total time= 0.0s
[CV 4/5] END .................................C=100, gamma=1; total time= 0.0s
[CV 5/5] END .................................C=100, gamma=1; total time= 0.0s
[CV 1/5] END ...............................C=100, gamma=0.1; total time= 0.0s
[CV 2/5] END ...............................C=100, gamma=0.1; total time= 0.0s
[CV 3/5] END ...............................C=100, gamma=0.1; total time= 0.0s
[CV 4/5] END ...............................C=100, gamma=0.1; total time= 0.0s
[CV 5/5] END ...............................C=100, gamma=0.1; total time= 0.0s
[CV 1/5] END ..............................C=100, gamma=0.01; total time= 0.0s
[CV 2/5] END ..............................C=100, gamma=0.01; total time= 0.0s
[CV 3/5] END ..............................C=100, gamma=0.01; total time= 0.0s
[CV 4/5] END ..............................C=100, gamma=0.01; total time= 0.0s
[CV 5/5] END ..............................C=100, gamma=0.01; total time= 0.0s
[CV 1/5] END .............................C=100, gamma=0.001; total time= 0.0s
[CV 2/5] END .............................C=100, gamma=0.001; total time= 0.0s
[CV 3/5] END .............................C=100, gamma=0.001; total time= 0.0s
[CV 4/5] END .............................C=100, gamma=0.001; total time= 0.0s
[CV 5/5] END .............................C=100, gamma=0.001; total time= 0.0s
[CV 1/5] END ............................C=100, gamma=0.0001; total time= 0.0s
[CV 2/5] END ............................C=100, gamma=0.0001; total time= 0.0s
[CV 3/5] END ............................C=100, gamma=0.0001; total time= 0.0s
[CV 4/5] END ............................C=100, gamma=0.0001; total time= 0.0s
[CV 5/5] END ............................C=100, gamma=0.0001; total time= 0.0s
[CV 1/5] END ................................C=1000, gamma=1; total time= 0.0s
[CV 2/5] END ................................C=1000, gamma=1; total time= 0.0s
[CV 3/5] END ................................C=1000, gamma=1; total time= 0.0s
[CV 4/5] END ................................C=1000, gamma=1; total time= 0.0s
[CV 5/5] END ................................C=1000, gamma=1; total time= 0.0s
[CV 1/5] END ..............................C=1000, gamma=0.1; total time= 0.0s
[CV 2/5] END ..............................C=1000, gamma=0.1; total time= 0.0s
[CV 3/5] END ..............................C=1000, gamma=0.1; total time= 0.0s
[CV 4/5] END ..............................C=1000, gamma=0.1; total time= 0.0s
[CV 5/5] END ..............................C=1000, gamma=0.1; total time= 0.0s
[CV 1/5] END .............................C=1000, gamma=0.01; total time= 0.0s
[CV 2/5] END .............................C=1000, gamma=0.01; total time= 0.0s
[CV 3/5] END .............................C=1000, gamma=0.01; total time= 0.0s
[CV 4/5] END .............................C=1000, gamma=0.01; total time= 0.0s
[CV 5/5] END .............................C=1000, gamma=0.01; total time= 0.0s
[CV 1/5] END ............................C=1000, gamma=0.001; total time= 0.0s
[CV 2/5] END ............................C=1000, gamma=0.001; total time= 0.0s
[CV 3/5] END ............................C=1000, gamma=0.001; total time= 0.0s
[CV 4/5] END ............................C=1000, gamma=0.001; total time= 0.0s
[CV 5/5] END ............................C=1000, gamma=0.001; total time= 0.0s
[CV 1/5] END ...........................C=1000, gamma=0.0001; total time= 0.0s
[CV 2/5] END ...........................C=1000, gamma=0.0001; total time= 0.0s
[CV 3/5] END ...........................C=1000, gamma=0.0001; total time= 0.0s
[CV 4/5] END ...........................C=1000, gamma=0.0001; total time= 0.0s
[CV 5/5] END ...........................C=1000, gamma=0.0001; total time= 0.0s